CATEGORY Publications

New publication!
Our new article has been accepted in Liver Cancer: Schotten C, Bechmann LP, Manka P, Theysohn J, Dechêne A, El Fouly A, Barbato F, Neumann U, Radünz S, Sydor S, Heider D, Venerito M, Canbay A, Gerken G, Herrmann K, Wedemeyer H, Best J: NAFLD-associated comorbidities in advanced stage HCC do not alter the safety…
READ MORE

New publication!
Our new article has been accepted in Bioinformatics: Löchel HF, Eger D, Sperlea T, Heider D: Deep Learning on Chaos Game Representation for Proteins. Bioinformatics 2019, in press. Abstract Motivation: Classification of protein sequences is one big task in bioinformatics and has many applications. Different machine learning methods exist and are applied on these problems,…
READ MORE

NETTAB/BBCC 2019
Dominik is member of the program commitee of the JOINT NETTAB/BBCC 2019 MEETING which will take place November 11-13, 2019 at the Fisciano Campus, University of Salerno, Italy http://www.nettab.org/2019/ https://www.bbcc-meetings.it/ DEADLINE FOR SUBMISSION OF ABSTRACTS FOR ORAL COMMUNICATIONS: Sunday June 30, 2019 TOPICS Oral presentations and posters are particularly encouraged for the usual topics of…
READ MORE

Talk at Harvard Medical School
Dominik gave a talk on Artificial Intelligence in Medicine at Harvard Medical School, Boston.
READ MORE

ISMB 2019
Anne-Christins abstract has been accepted at the ISMB 2019 and she will present a poster on “ The Importance of Privacy Preserving Machine Learning in Psychiatric Research„ Looking forward to ISMB 2019 in Basel!
READ MORE

Talks at GCB 2019
Franzi’s and Theo’s abstracts have been accepted at the GCB 2019 and they will give talks on Deep Learning in Chaos Game Representation for Proteins and On the semantics of unknown DNA motifs – a machine learning approach respectively. Looking forward to GCB 2019 in Heidelberg!
READ MORE

Talk at Rutgers University
Dominik gave a talk on Biomedical Big Data Science at the Department of Microbiology at Rutgers University.
READ MORE
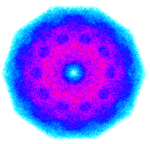
Chaos Game Representation
We recently developed an R package kaos, which can be used to build fractals from sequences of symbols. These fractals are based on the Chaos Game Representation (CGR), which is a recurrent iterative function system. While CGR has been used mainly for genome sequence encoding and classification in the context of bioinformatics, we modified it…
READ MORE

New publication!
Our paper has been accepted in PLoS One: Canbay A, Kälsch J, Neumann U, Rau M, Hohenester S, Strucksberg K-H, Baba HA, Rust C, Bantel H, Geier A, Heider D, Sowa J-P: Non-invasive assessment of NAFLD as systemic disease – a machine learning perspective. PLoS One 2019, 14(3):e0214436. Abstract Background & aims Current non-invasive scores…
READ MORE

New publication!
Our paper has been accepted in BioData Mining: Spänig S, Heider D: Encodings and models for antimicrobial peptide classification for multi-resistant pathogens. BioData Mining 2019, 12:7. (Link) Abstract Antimicrobial peptides (AMPs) are part of the inherent immune system. In fact, they occur in almost all organisms including, e.g., plants, animals, and humans. Remarkably, they show…
READ MORE







